| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| 19 | 20 | 21 | 22 | 23 | 24 | 25 |
| 26 | 27 | 28 | 29 | 30 | 31 |
Tags
- 다변량분석
- 서평
- 소셜네트워크분석
- cnn
- 에이블스쿨
- 한국전자통신연구원 인턴
- python
- Eda
- 딥러닝
- hadoop
- ML
- httr
- 시계열
- 머신러닝
- matplot
- 하둡
- 하계인턴
- 지도학습
- dx
- 시각화
- KT 에이블스쿨
- ggplot2
- 빅분기
- 한국전자통신연구원
- r
- 웹크롤링
- 기계학습
- arima
- KT AIVLE
- 가나다영
- ETRI
- 빅데이터분석기사
- SQL
- kaggle
- 에트리 인턴
- Ai
- SQLD
- 에이블러
- kt aivle school
- 프로그래머스
Archives
- Today
- Total
소품집
[다변량 분석] 캐글 Mushrooms Data Classification 본문
728x90
setwd('/Users/dayeong/Desktop/21-2/전공/다변량 분석')
# Pakage
library(dplyr)
library(ggplot2)
library(caret)
library(rpart)
library(rpart.plot)
library(randomForest)
# Data Set roding
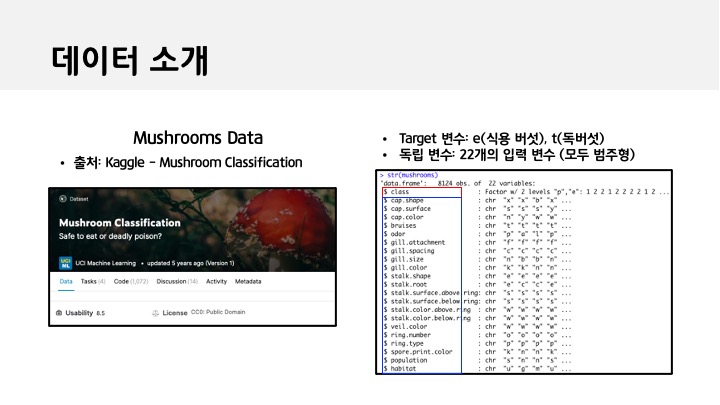
mushrooms <- read.csv('mushrooms.csv')
for (i in 2:23) {
test <- chisq.test(table(mushrooms$class, mushrooms[,i]))
if (test$p.value < 0.05) {
print(test)
}
}
# Target 변수 확인
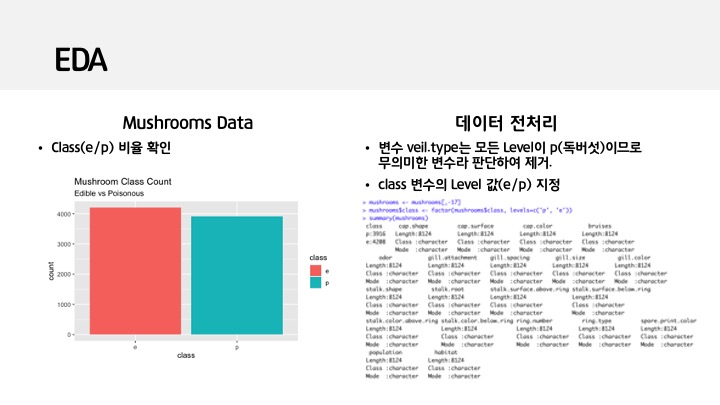
ggplot(data=mushrooms, aes(x=class, fill=class)) +
geom_bar()+
labs(title='Mushroom Class Count',subtitle = 'Edible vs Poisonous')
# veil.type 변수는 모두 p(poisonus) -level이 1인 변수로 무의미하다 판단하여 제거.
mushrooms <- mushrooms[,-17]
mushrooms$class <- factor(mushrooms$class, levels=c('p', 'e'))
summary(mushrooms)
# Train / Test set Split
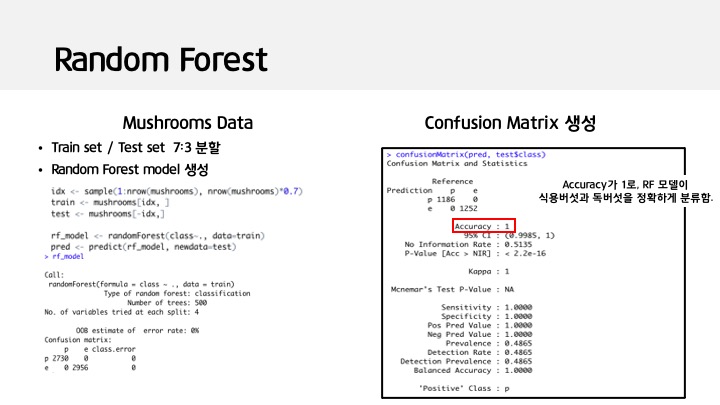
idx <- sample(1:nrow(mushrooms), nrow(mushrooms)*0.7)
train <- mushrooms[idx, ]
test <- mushrooms[-idx,]
# RandomForest
rf_model <- randomForest(class~., data=train)
pred <- predict(rf_model, newdata=test)
confusionMatrix(pred, test$class)
# Decision Tree
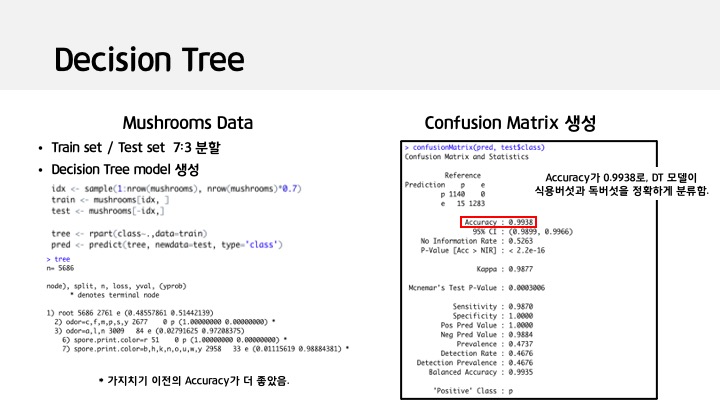
idx <- sample(1:nrow(mushrooms), nrow(mushrooms)*0.7)
train <- mushrooms[idx, ]
test <- mushrooms[-idx,]
tree <- rpart(class~.,data=train)
summary(tree)
pred <- predict(tree, newdata=test, type='class')
confusionMatrix(pred, test$class)
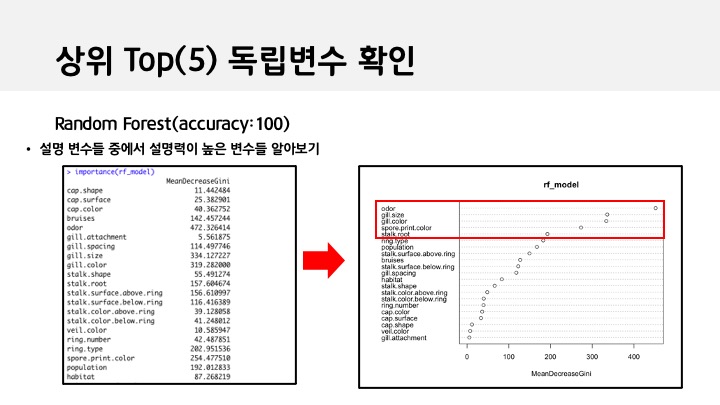
importance(rf_model)
importance(tree)
varImpPlot(rf_model)
varImpPlot(tree)
test$class <- predict(tree, test, type='class')
test$pred <- pred
ggplot(data=test, aes(class, pred)) +
geom_jitter(width = 0.2, height = 0.1, size=2)(HW02)다변량분석-20181478 소다영.pdf
1.39MB



728x90
'Statistics' 카테고리의 다른 글
| [다변량분석] 회귀분석 - Prestige Data 잔차 분석, 모델 성능 비교 (0) | 2022.12.14 |
|---|---|
| [다변량 분석] 모형 적합성, 회귀계수 유의성 검정, 결정계수(R^2) 해석 (0) | 2021.10.13 |
| [다변량 분석] Survey Data를 이용한 다변량분석 (0) | 2021.09.29 |
| [다변량 분석] 검정 및 신뢰구간 추정 (1) | 2021.09.29 |
| statistics (2) | 2020.12.05 |
Comments


